Downoading data from PolicyMap
CRD 230 - Spatial Methods in Community Research
Professor Noli Brazil
This guide provides step-by-step instructions for downloading data from PolicyMap. PolicyMap is a fully web-based online data and mapping application that gives you access to over 15,000 indicators related to demographics, housing, crime, mortgages, health, jobs and more. Data are available at all common geographies (address, block group, census tract, zip code, county, city, state, Metropolitan area) as well as unique geographies like school districts and political boundaries.
In this guide, we will download PolicyMap census tract data for the City of Oakland. We will download median housing value. UC Davis provides full access to all PolicyMap tools for staff, students, and faculty. You can access the full site only if you are on campus or logged onto the UC Davis VPN Client. Download the PulseSecure VPN app using the directions outlined here. We will download data from the UCD PolicyMap portal and clean up the files in R.
Installing and Loading Packages
We’ll be using the package VIM in this guide. Install it.
install.packages("VIM")Then load it and the other packages we will be using in this guide.
library(tidyverse)
library(tidycensus)
library(VIM)Download data
- Navigate to the UC Davis PolicyMap portal. You should see a webpage that looks like the figure below. Note the UC Davis logo on the top right. Go Aggies!
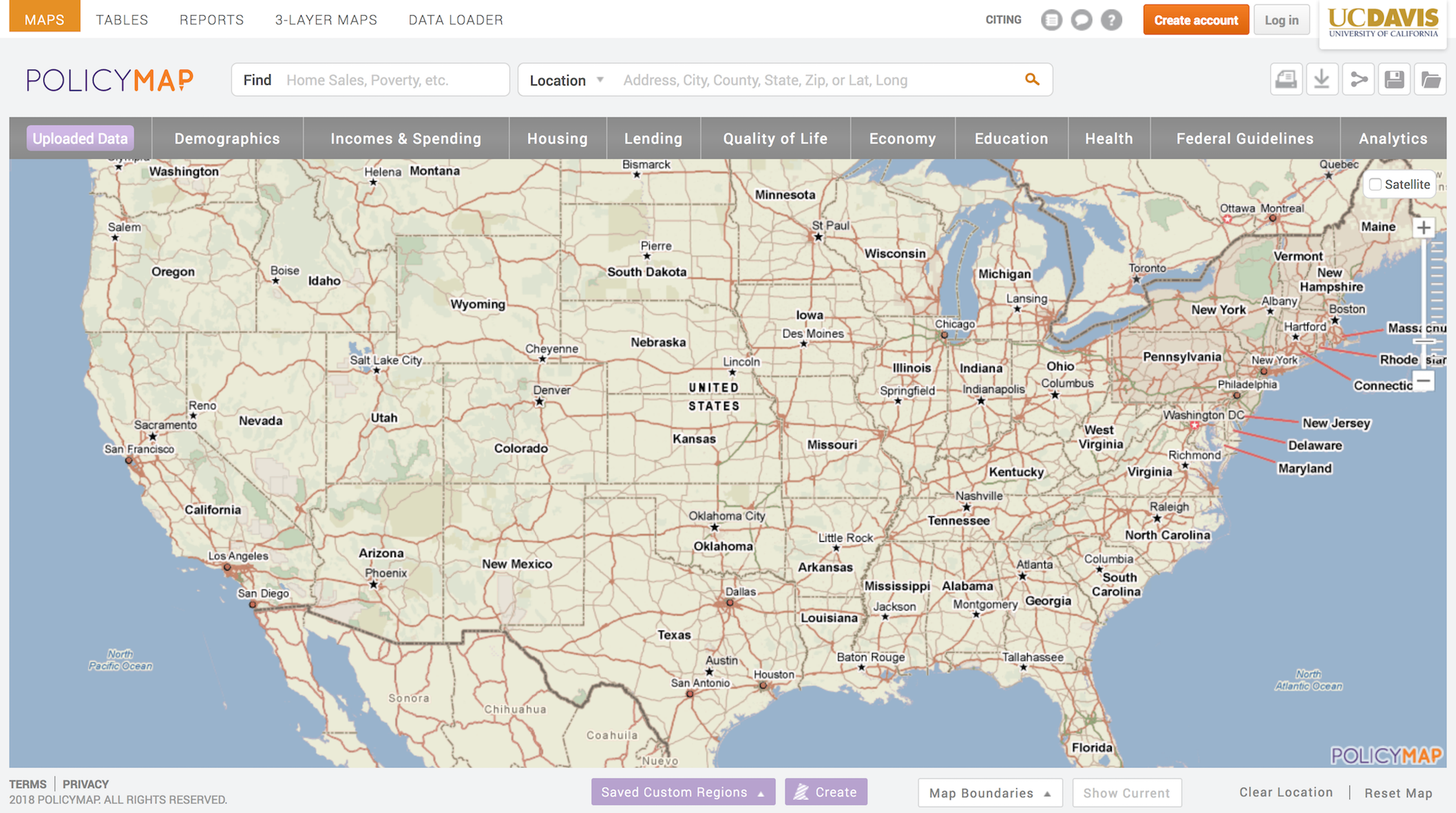
- You should see a Location search bar somewhere near the top of the page. Type in “Oakland, CA” in the search bar and Oakland, CA (City) should pop up below - select it.
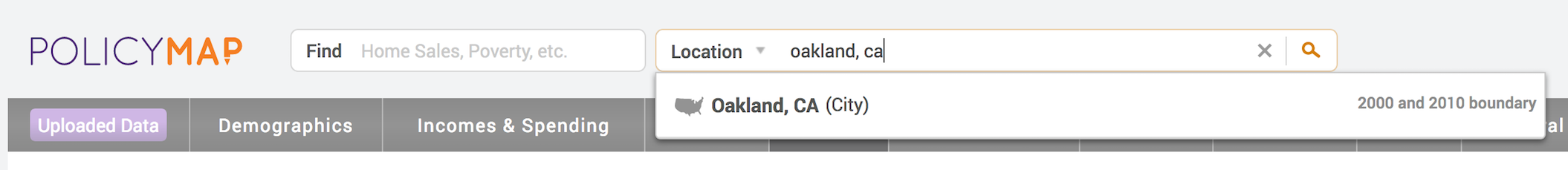
You should get a map that highlights Oakland’s boundaries.
- The map does not show any data. Let’s add the Median Housing Value. Click on the Housing tab, followed by Median Value under Home Values.
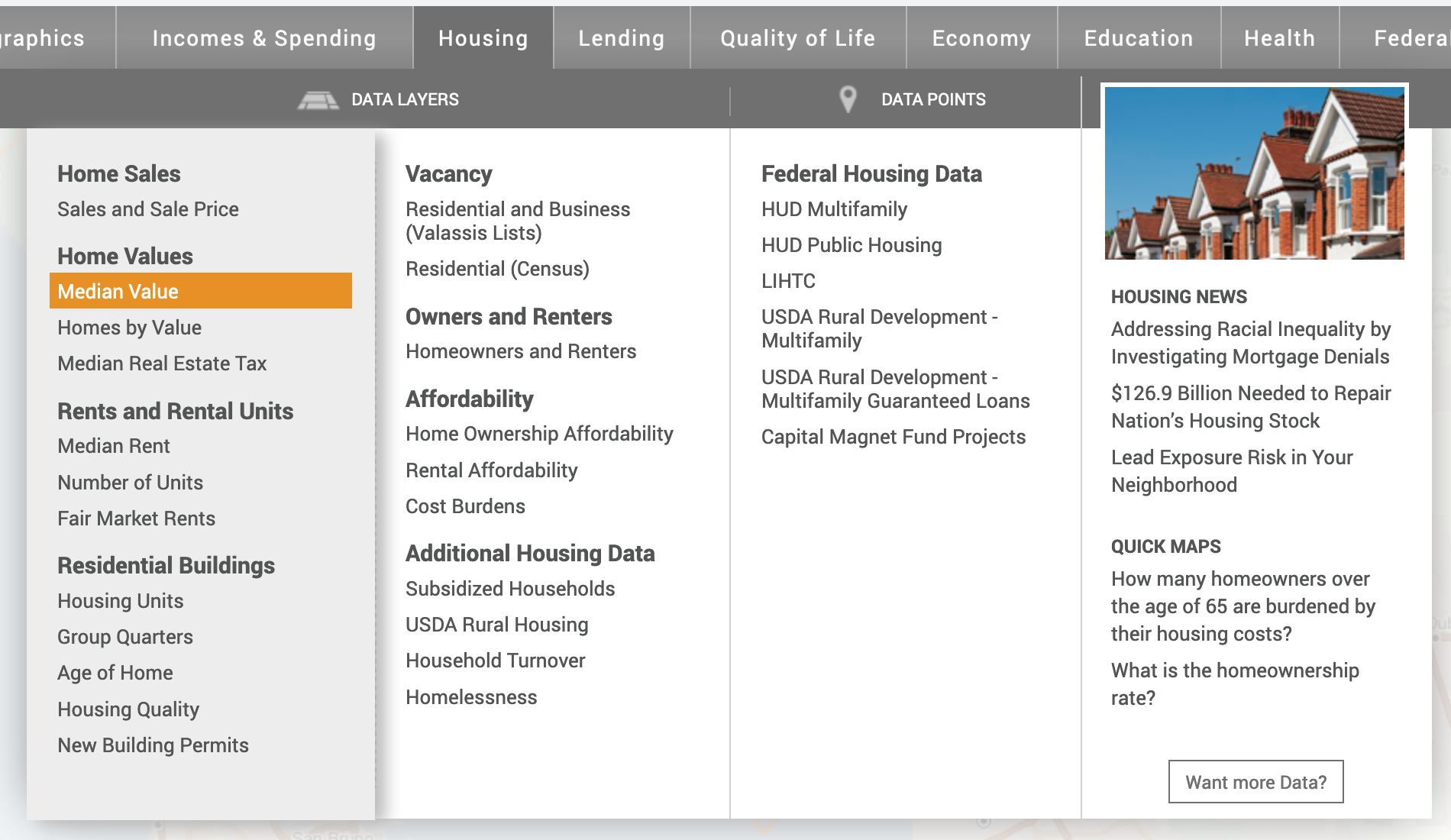
Now your map should look like the following
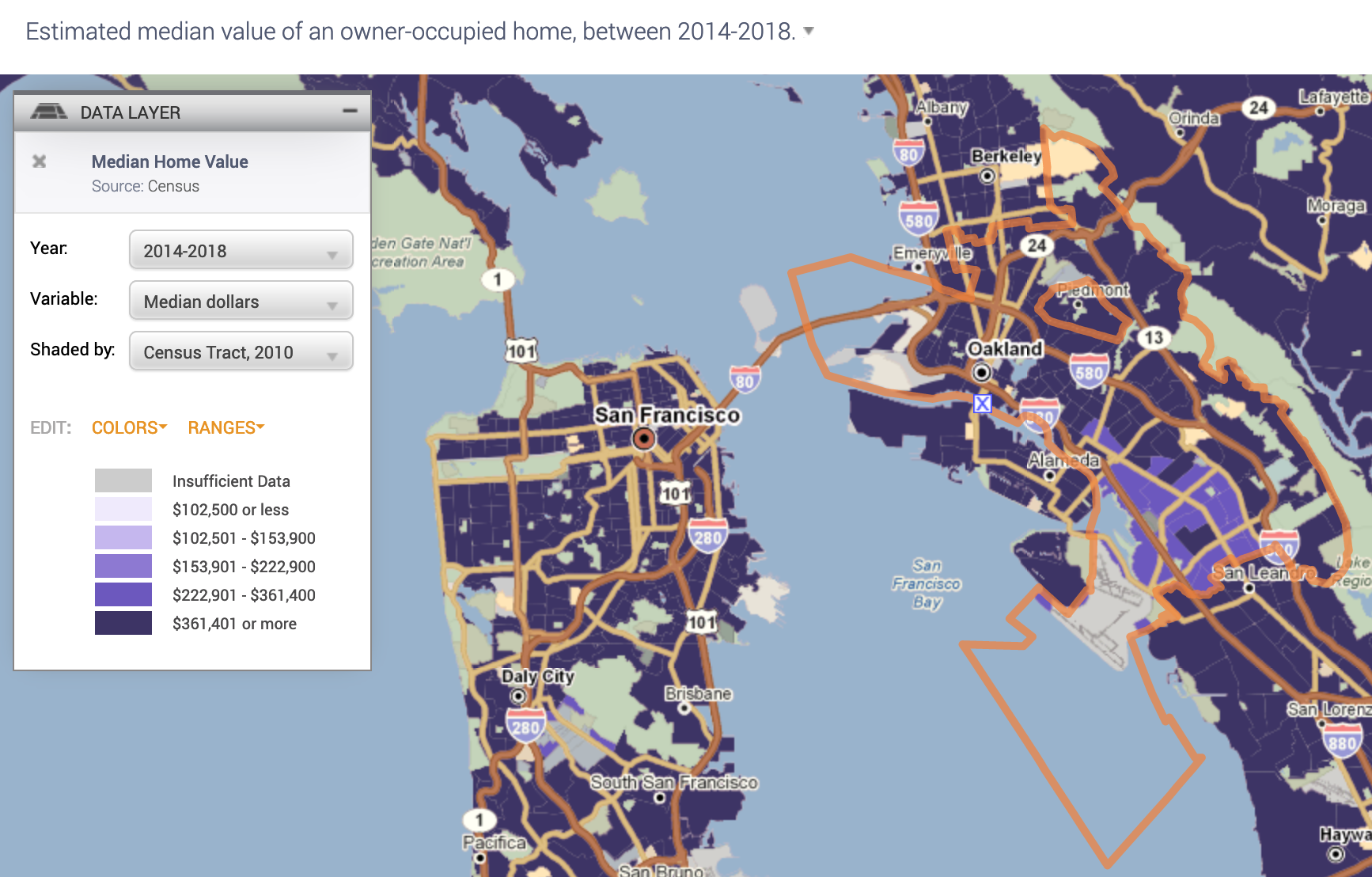
- Notice in the legend window you can change various aspects of the variable, including the year
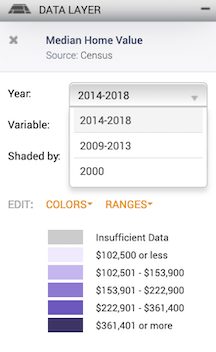
the data type
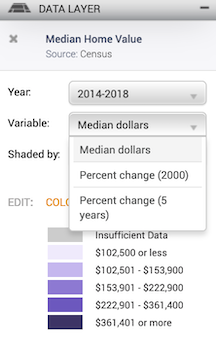
and the geographic level.
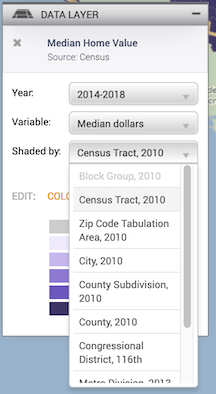
Leave the defaults (Year: 2014-2018, Variable: Median dollars, and Shaded by: Census Tract, 2010).
Let’s download these data. At the top right of the site, click on the download icon
 .
.A window should pop up. The first screen asks you what data to download - it should be “Estimated median value of an owner-occupied home, between 2014-2018” under Layer. Click on the Next, Select Location button
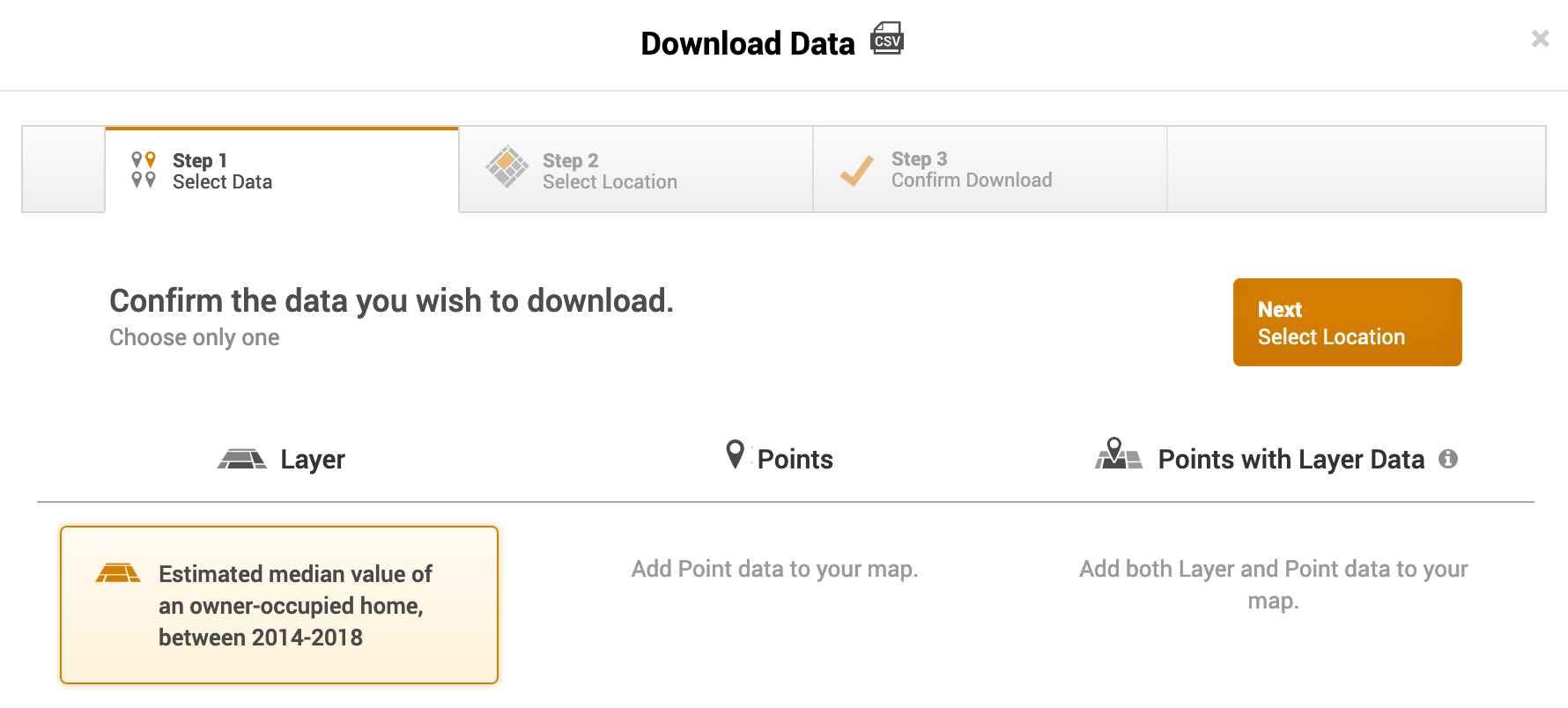
- The next screen asks you to select a location. It should be Oakland City, CA (City, 2010) and Shaded by Census Tract. Click on Next, Confirm Download.
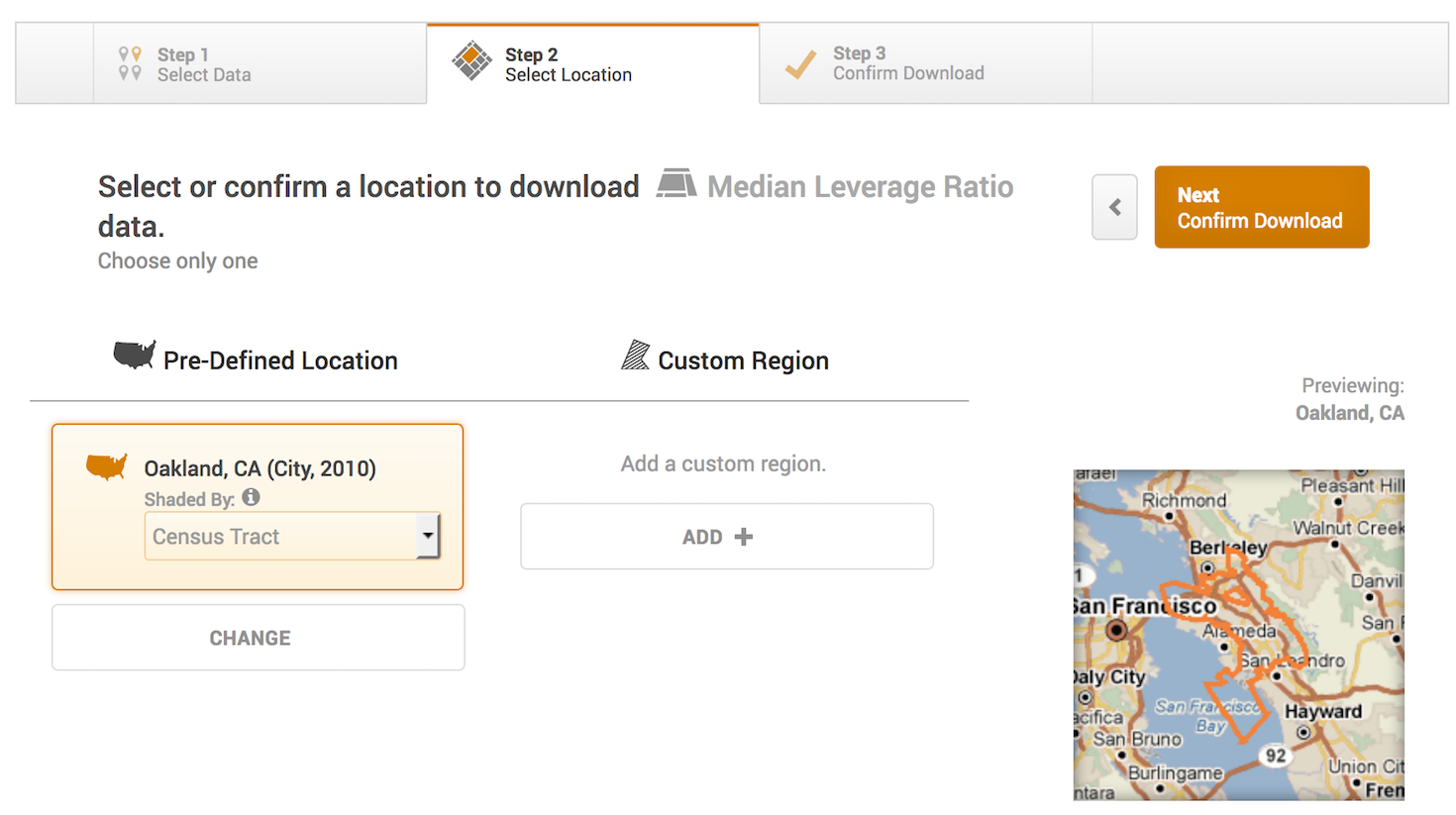
The next screen asks you to confirm the download - just click on Download CSV

After a minute or two, a screen like below (on a Mac laptop) should pop up (the file name will differ).
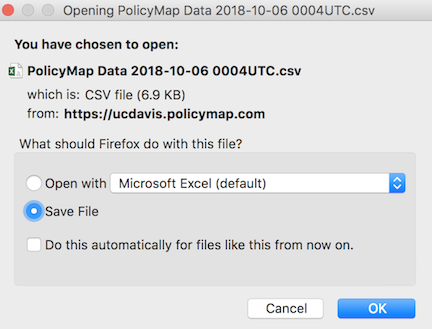
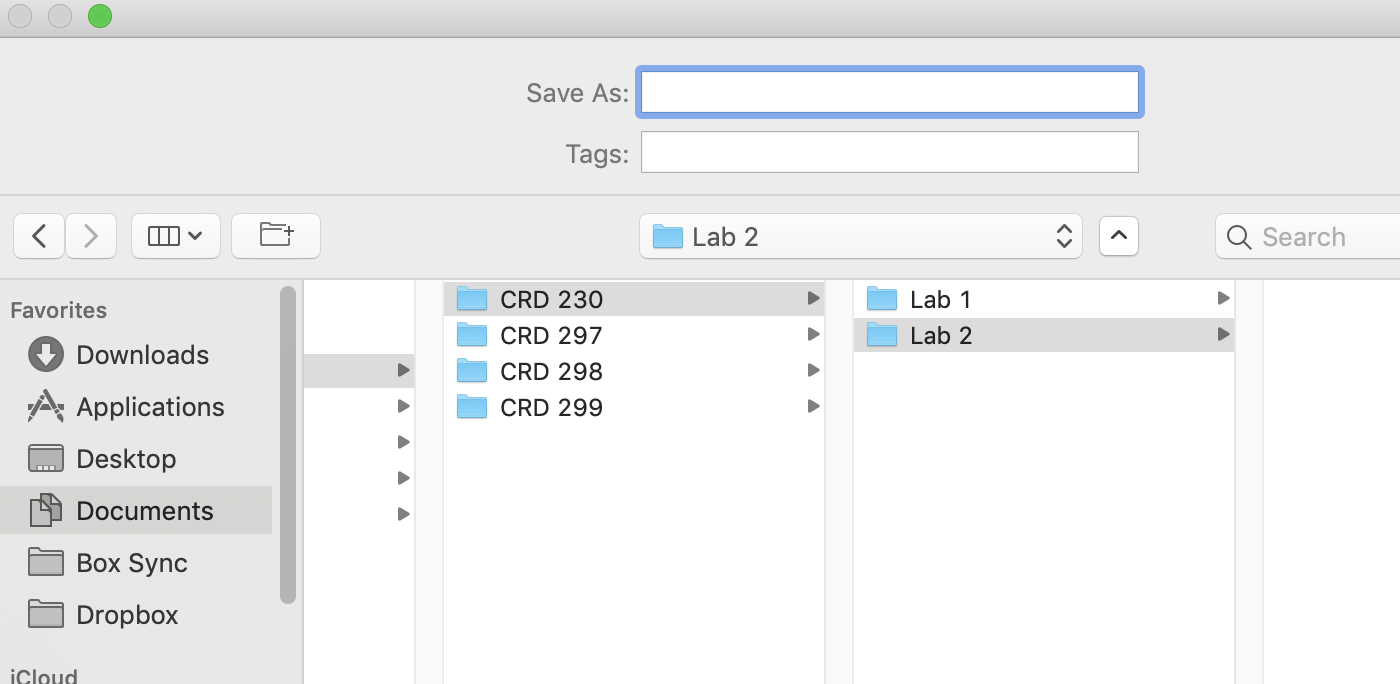
Save the file into an appropriate folder, such as below (Mac laptop)
- PolicyMap allows you to download only one variable at the time. So, you’ll need to go through the above steps again to get other variables. Follow the same steps above to download these data as a csv into an appropriate folder on your hard drive. For example, to get percent black, navigate to the Demographics tab, then under Race select Black. To download percent Hispanic, navigate to the Demographics tab, then under Ethnicity select Hispanic and finally All.
Data Wrangling in R
Bring in the data using read_csv().
pm.file <- read_csv("##YOUR FILE NAME HERE")We need to clean up the file to get it ready for analysis. The first issue with pm.file is the footnotes located at the bottom of the file. If you view the file in R, the footnotes look like
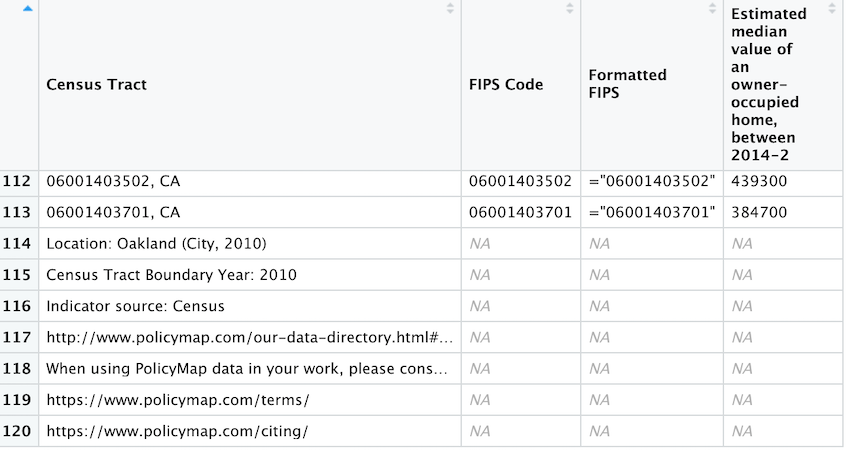
To remove the footnotes, use the function slice()
pm.file <- pm.file %>%
slice(-(114:n()))The function removes all rows starting from row 114 to the last row in the tibble (last row is indicated by the function n()). Note the negative sign that tells the function to remove the rows. Without the negative sign, the function will instead keep those rows.
We also need to rename variables and keep the ones that are relevant. First, let’s look at the pm.file’s column names
names(pm.file)## [1] "Census Tract"
## [2] "FIPS Code"
## [3] "Formatted FIPS"
## [4] "Estimated median value of an owner-occupied home, between 2014-2"The variable Estimated median value of an owner-occupied home, between 2014-2 contains median housing values. Let’s shorten the name because we would not want to have to type this long name out everytime we want to refer to this variable. Make the name simple and clear. Here, we rename it medval using the rename() function.
pm.file <- pm.file %>%
rename(medval = "Estimated median value of an owner-occupied home, between 2014-2")We had to put quotes around the original variable name because it had spaces in between.
Next, let’s keep the necessary variables: FIPS Code and medinc. FIPS Code is the tract GEOID.
pm.file <- pm.file %>%
select("FIPS Code", medval)Merging with Census data
Let’s bring in some census data from the Census API. We covered how to use get_acs() from the tidycensus package in Lab 2.
ca <- get_acs(geography = "tract",
year = 2018,
variables = c(tpopr = "B03002_001",
nhwhite = "B03002_003", nhblk = "B03002_004",
nhasn = "B03002_006", hisp = "B03002_012"),
state = "CA",
survey = "acs5")
ca <- ca %>%
select(-moe) %>%
spread(key = variable, value = estimate)Merge in ca into pm.file. The linking variable is FIPS Code in pm.file and GEOID in ca.
pm.file <- pm.file %>%
left_join(ca, by = c("FIPS Code" = "GEOID"))Dealing with missing values
Missing values are a part of a social scientist’s life. You can’t avoid them.
You will notice that PolicyMap designates missing values as N/A. R designates missing as NA. Therefore, R reads N/A as a character. This means that R does not recognize medval as a numeric but a character.
class(pm.file$medval)## [1] "character"We need to replace N/A with NA and then convert it to numeric
pm.file <- pm.file %>%
mutate(medval = as.numeric(ifelse(medval == "N/A", "NA", medval)))The code ifelse(medval == "N/A", "NA", medval) says that if the variable medval equals “N/A”, replace it to “NA”, otherwise keep it its original value stored in medval. as.numeric() converts medval to a numeric.
class(pm.file$medval)## [1] "numeric"The next step is to determine what percentage of your cases are missing data. The best function for doing this in R is aggr(), which is in the VIM package. Run the aggr() function as follows
summary(aggr(pm.file))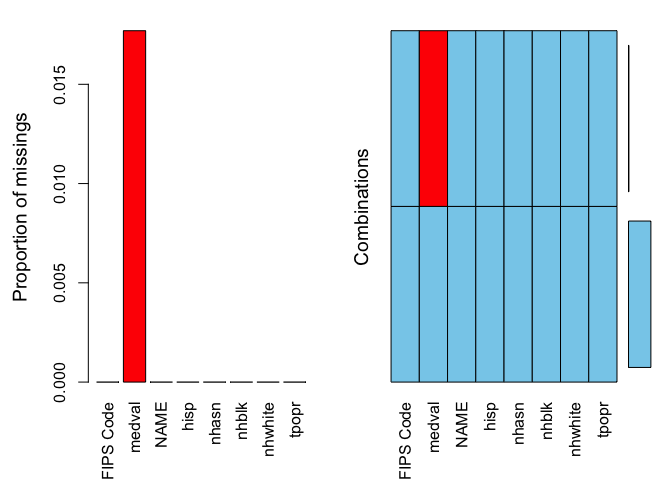
##
## Missings per variable:
## Variable Count
## FIPS Code 0
## medval 2
## NAME 0
## hisp 0
## nhasn 0
## nhblk 0
## nhwhite 0
## tpopr 0
##
## Missings in combinations of variables:
## Combinations Count Percent
## 0:0:0:0:0:0:0:0 111 98.230088
## 0:1:0:0:0:0:0:0 2 1.769912The results show two tables and two plots. The left-hand side plot shows the proportion of cases that are missing values for each variable in the data set. The right-hand side plot shows which combinations of variables are missing. The first table shows the number of cases that are missing values for each variable in the data set. The second table shows the percent of cases missing values based on combinations of variables. The results show that 2 or 1.8% of census tracts are missing values on the variable medval.
In any statistical analysis, you will need to deal with missing values. For example, if you wanted to find out the average median housing value in Oakland tracts, you would type in
mean(pm.file$medval)## [1] NAThe mean is NA, which tells you that there are missing values in the variable medval that you need to deal with before R calculates a value. There are many ways that one can deal with missing data. One method is to just simply ignore or discard cases with a missing value. To do this in the mean() function (and in many other R functions), you include the argument na.rm = TRUE
mean(pm.file$medval, na.rm = TRUE)## [1] 589359.5As long as a large proportion of your data set is not missing data, simply ignoring missing data is often acceptable. Just make sure you are transparent about what you did.

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Website created and maintained by Noli Brazil